Table Of Content
- Design of highly functional genome editors by modeling the universe of CRISPR-Cas sequences
- Computer-based design of sensors that can monitor endogenous Ras activity
- Functional-motif scaffolding
- Membrane proteins
- Design of globular proteins
- Designing hydrogels with the DeForest Lab
- A computationally designed chimeric antigen receptor provides a small-molecule safety switch for T-cell therapy

“A regional hub is a very natural thing to emerge,” said Portland, Ore.-based Jonathan Cohen, vice president of applied research at NVIDIA, which is investing heavily in AI-mediated drug design. Research on it began here as an undergraduate project and would go on to form the basis of a spinout company. Advanced Drug DeliveryNanoscale protein assemblies that move therapeutics to specific cells within the body. They are found inside every living thing and act as structural components, transporters, signaling molecules, and much more.
Design of highly functional genome editors by modeling the universe of CRISPR-Cas sequences
The same feat can be achieved with protein sequences and structures, where the algorithm draws on a rich repository of real-world biological information to dream up new proteins based on the patterns and principles observed in nature. To do this, however, researchers also need to give the computer guidance on the biochemical and physical constraints that inform protein design, or else the resulting output will offer little more than artistic value. A different strategy incorporates human expert knowledge into the process of backbone generation for design. The TopoBuilder (72) protocol lets designers build proteins in a bottom-up approach starting from functional motifs (e.g., a helix in a binding interface). Designers define the sizes and three-dimensional coordinates of secondary structure elements.
Computer-based design of sensors that can monitor endogenous Ras activity
Both max-product and sum-product belief propagation have been used to optimize protein design. Other powerful extensions to the dead-end elimination algorithm include the pairs elimination criterion, and the generalized dead-end elimination criterion. This algorithm has also been extended to handle continuous rotamers with provable guarantees. The IPD generates open-source AI tools to craft protein-based therapeutics, vaccines, materials and biosensors, and its Seattle-area spinouts and affiliated companies interact with each other and partner with larger biopharma companies. If you want an amazing protein label that stands out from the competition, work with a professional designer.
Functional-motif scaffolding
“They are models, so you have to take them with a grain of salt, but now you have this extraordinarily large amount of predicted structures that you can build upon,” says Zanghellini, who says this tool is a core component of Arzeda’s computational design workflow. One effective strategy to understand protein sequence and structure is to approach them as ‘text’, using language modeling algorithms that follow rules of biological ‘grammar’ and ‘syntax’. In a recent publication, Madani and colleagues describe a language modeling algorithm that can yield novel computer-designed proteins that can be successfully produced in the lab with catalytic activities comparable to those of natural enzymes. Language modeling is also a key part of Arzeda’s toolbox, according to co-founder and CEO Alexandre Zanghellini. For one project, the company used multiple rounds of algorithmic design and optimization to engineer an enzyme with improved stability against degradation.
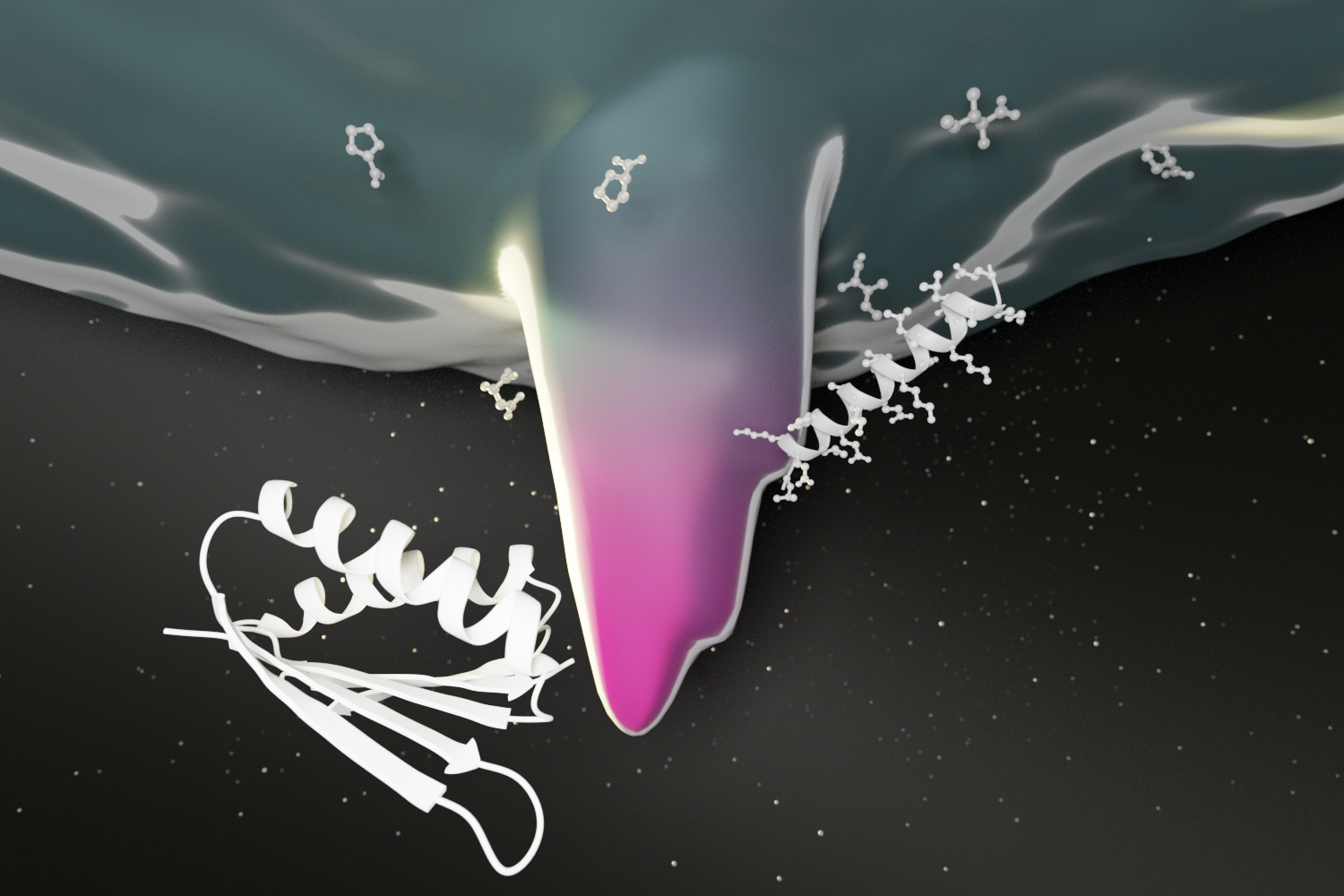
Membrane proteins
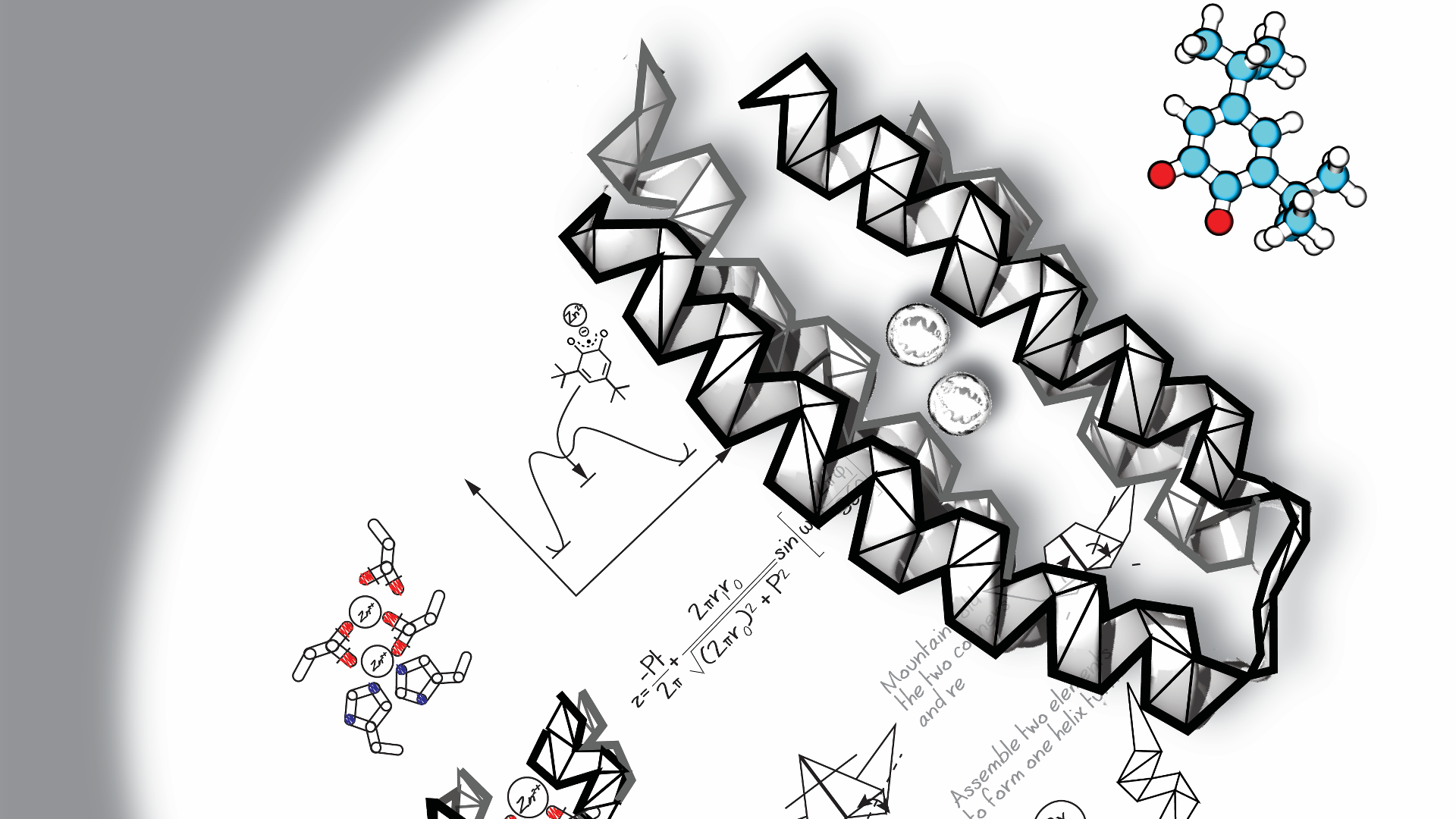
Symmetry was used to design a 4-fold (colors) symmetric TIM barrel (34) and repeat proteins (67). D, de novo protein fold families can be generated by sampling the geometries (length, as well as relative position and orientation) of secondary structure elements (28, 32). E, generative machine learning methods (red) build novel backbone structures by latent space sampling (81). The hallucination method (45) (red) uses the TR-Rosetta neural network to predict the structure distribution of a sequence. The sequence is optimized using Monte Carlo–simulated annealing by maximizing the divergence between the predicted structure distribution and a background distribution representing unstructured proteins.
Design of globular proteins
Tandem repeat proteins made of a series of identical helix–loop–helix–loop structural motifs can be systematically assembled (67). By modulating the curvature, alpha tandem repeat proteins can form closed toroid structures (68). A large number of proteins with diverse shapes can be generated by designing rigid junctions to connect helical repeat proteins (69).
The global protein design and engineering market has advanced due in large part to advancements in genetic engineering techniques. Scientists now have amazing tools at their disposal for designing and producing novel proteins with enhanced features and capabilities thanks to directed evolution and recombinant DNA technologies. The high demand for tailored medications, technological improvements, and the growing incidence of infectious, chronic, and protein-deficient disorders are some of the drivers propelling the global market for protein design and engineering. Moreover, the expansion of the global industry can also be attributed to the government's increased support for protein engineering R&D, the availability of reliable healthcare infrastructures, and advancements in bioinformatics databases and tools.
The UW's Institute for Protein Design keeps boosting startups, fueling AI-powered science - GeekWire
The UW's Institute for Protein Design keeps boosting startups, fueling AI-powered science.
Posted: Thu, 25 Apr 2024 19:35:27 GMT [source]
A computationally designed chimeric antigen receptor provides a small-molecule safety switch for T-cell therapy
Challenges are particularly apparent in the design of proteins with new functions (Fig. 7). New protein structures can be designed with considerable success rates without experimental optimization (Table 1), but the activities of proteins derived directly from the computational design are often weaker than achievable activities of naturally evolved proteins. Therefore, computational designs are often (although not always) optimized by experimental methods such as site saturation mutagenesis (4, 20). The de novo ligand-binding site design requires high accuracy in sampling and scoring.
In particular, AbDesign breaks proteins from a structure family into a few modular segments based on structural alignments and then recombines these segments into new backbones. AbDesign is able to build large numbers of similar structures even for moderately sized families of homologs. An elegant example of a protein design approach to delineate protein function in vivo is seen in the work of Shokat et al.73 By designing a protein kinase with altered ATP-binding specificity they were able to identify substrates of that kinase. The strategy was to first introduce mutations in the kinase that enlarged the substrate-binding pocket such that bulky ATP derivatives could be bound. The binding pocket on the wild-type kinase is too small to bind the bulky ATP derivative.
Self-Assembling NanomaterialsAtomically precise materials with applications in solar energy, imaging, and basic research. Reprinted with permission from Tsien, R. Y. Angewandte Chemie International Edition 2009, 48, 5612–5626. All rights are reserved, including those for text and data mining, AI training, and similar technologies. The world of proteins is incredibly complex, offering endless possibilities for exploration and discovery.
Assessing the laboratory performance of AI-generated enzymes - Nature.com
Assessing the laboratory performance of AI-generated enzymes.
Posted: Tue, 23 Apr 2024 09:33:32 GMT [source]
In the following sections, we describe advances in the design in the areas of binding proteins for ligands and other proteins, large protein assemblies, membrane proteins, and protein switches. Solutions of fixed backbone side-chain design problems are sensitive to the backbone structures used as input. Because the Lennard-Jones potential term in scoring functions (see the section below) scales as the 12th power of distance when two atoms are close to each other, a small adjustment to the backbone structure may result in a considerable energy change.
Another crucial component of the design is the orthogonality of the pairs—unintentional cross-reactivity between different coiled-coil monomers would prevent the proper assembly of the tetrahedron. The resulting 3D structure was imaged by atomic force microscopy (AFM), and the proximity of the N- and C-termini at the same vertex was confirmed by a split-fluorescent protein assay. Protein origami is attractive because such structures can be easily functionalized for use in pathway engineering, difference imaging, and novel vaccines. However, a rational protein design approach must model some flexibility on the target structure in order to increase the number of sequences that can be designed for that structure and to minimize the chance of a sequence folding to a different structure. For example, in a protein redesign of one small amino acid (such as alanine) in the tightly packed core of a protein, very few mutants would be predicted by a rational design approach to fold to the target structure, if the surrounding side-chains are not allowed to be repacked. The success rate is defined as the percentage of reported designs in each study that adopt the designed structure (folded, blue; experimental structure determined, orange) or function (green, red).
In a manner analogous to networks that produce images from user-specified inputs, RFdiffusion enables the design of diverse functional proteins from simple molecular specifications. A number of machine learning methods for protein sequence design were developed recently (Fig. 3E). Deep neural network methods were trained to predict probabilities of amino acids at each residue position of a backbone structure (102, 103). Generative models learn distributions of protein sequences and can generate new native-like protein sequences with or without input backbone structures. A number of generative models were developed for sequence design, including generative adversarial networks (104), variational autoencoders (105, 106), and graph-based (107, 108) models.
No comments:
Post a Comment